Science
Deep Learning Transforms 3D Imaging of Fruit Microstructure
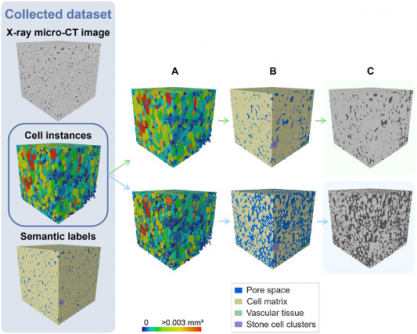
Recent advancements in deep learning have revolutionized the study of plant tissue microstructure, particularly in apples and pears. A research team led by Pieter Verboven from KU Leuven has developed a novel framework that automates the labeling and quantification of plant tissues using X-ray micro-computed tomography (micro-CT). Published in Plant Phenomics on July 5, 2025, this work marks a significant leap over traditional two-dimensional imaging techniques, achieving high accuracy in three-dimensional analysis.
The structure of plant tissues plays a critical role in their metabolic processes, yet conventional microscopy methods often require extensive sample preparation and provide limited views. Traditional imaging techniques struggle with the complexity of overlapping features and low image contrast, making it challenging to accurately quantify tissue morphology. While X-ray micro-CT has facilitated non-destructive imaging, the segmentation of various cell types has remained difficult.
To tackle these issues, the research team employed a 3D panoptic segmentation framework that builds on the existing 3D extension of Cellpose and a 3D Residual U-Net architecture. This model allows for comprehensive labeling of fruit tissue microstructures directly from micro-CT images. The framework executes both instance segmentation—identifying separate parenchyma cells—and semantic segmentation, which classifies voxels into distinct categories such as pore space and vascular structures.
Training the model involved utilizing datasets from apples and pears, supplemented with synthetic data augmentation techniques that included morphological dilation and erosion, grey-value assignment, and the addition of Gaussian noise. The model was rigorously compared against a two-dimensional instance segmentation model and a marker-based watershed algorithm. Notably, the 3D model achieved an Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears. In contrast, the 2D model recorded AJIs of 0.861 for apples and 0.732 for pears.
Evaluation metrics demonstrated that the model accurately segmented pore spaces and cell matrices while effectively identifying vascular tissues and stone cell clusters. For instance, the Dice Similarity Coefficient (DSC) indicated a score of 0.506 for apple vasculature and 0.789 for pear vasculature. Visual validation confirmed the model’s efficacy in detecting vascular bundles in apple varieties such as ‘Kizuri’ and ‘Braeburn’, as well as realistic segmentation of stone cell clusters in pear varieties like ‘Celina’ and ‘Fred’.
Despite its success, the study acknowledged limitations related to dataset imbalance and domain shifts, which hindered further performance enhancements through additional data augmentation. Morphometric analysis affirmed the model’s reliability, revealing vasculature widths ranging from 70 to 780 μm and variability in stone cell cluster dimensions and sphericity.
The implications of this research extend far beyond basic tissue analysis. The deep learning model represents a powerful, non-destructive tool for plant scientists, facilitating studies on how microscopic structures influence essential processes such as water, gas, and nutrient transport. By significantly reducing the manual effort required for tissue characterization, the model accelerates what is often a labor-intensive analysis.
In the realm of fruit research, understanding cellular arrangements is crucial for determining textural qualities, storability, and susceptibility to physiological disorders like browning or watercore. Furthermore, this technology offers a scalable solution for examining tissue development, ripening, and stress responses across various crops. Its compatibility with standard X-ray micro-CT instruments positions it as an accessible resource for integrating artificial intelligence into the fields of plant anatomy and food science.
This research was supported by the Research Foundation – Flanders (FWO) under grant number S003421N, as part of the SBO project FoodPhase, alongside contributions from KU Leuven.
In summary, the 3D deep learning model developed by Verboven’s team heralds a new era in plant tissue analysis, promising greater accuracy and efficiency in studies essential for advancing agricultural science and food quality research.
-
 Science2 months ago
Science2 months agoOhio State Study Uncovers Brain Connectivity and Function Links
-

 Politics2 months ago
Politics2 months agoHamas Chief Stresses Disarmament Tied to Occupation’s End
-

 Science1 month ago
Science1 month agoUniversity of Hawaiʻi Joins $25.6M AI Project for Disaster Monitoring
-
 Science4 weeks ago
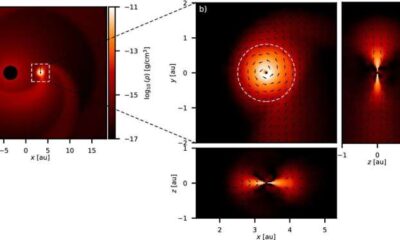
Science4 weeks agoALMA Discovers Companion Orbiting Giant Star π 1 Gruis
-

 Entertainment2 months ago
Entertainment2 months agoMegan Thee Stallion Exposes Alleged Online Attack by Bots
-

 Science2 months ago
Science2 months agoResearchers Challenge 200-Year-Old Physics Principle with Atomic Engines
-

 Entertainment2 months ago
Entertainment2 months agoPaloma Elsesser Shines at LA Event with Iconic Slicked-Back Bun
-

 World1 month ago
World1 month agoFDA Unveils Plan to Cut Drug Prices and Boost Biosimilars
-

 Business2 months ago
Business2 months agoMotley Fool Wealth Management Reduces Medtronic Holdings by 14.7%
-

 Science2 months ago
Science2 months agoInnovator Captures Light at 2 Billion Frames Per Second
-

 Top Stories2 months ago
Top Stories2 months agoFederal Agents Detain Driver in Addison; Protests Erupt Immediately
-

 Entertainment1 month ago
Entertainment1 month agoBeloved Artist and Community Leader Gloria Rosencrants Passes Away









