Science
Researchers Unveil Groundbreaking Chromatin Mapping Technique
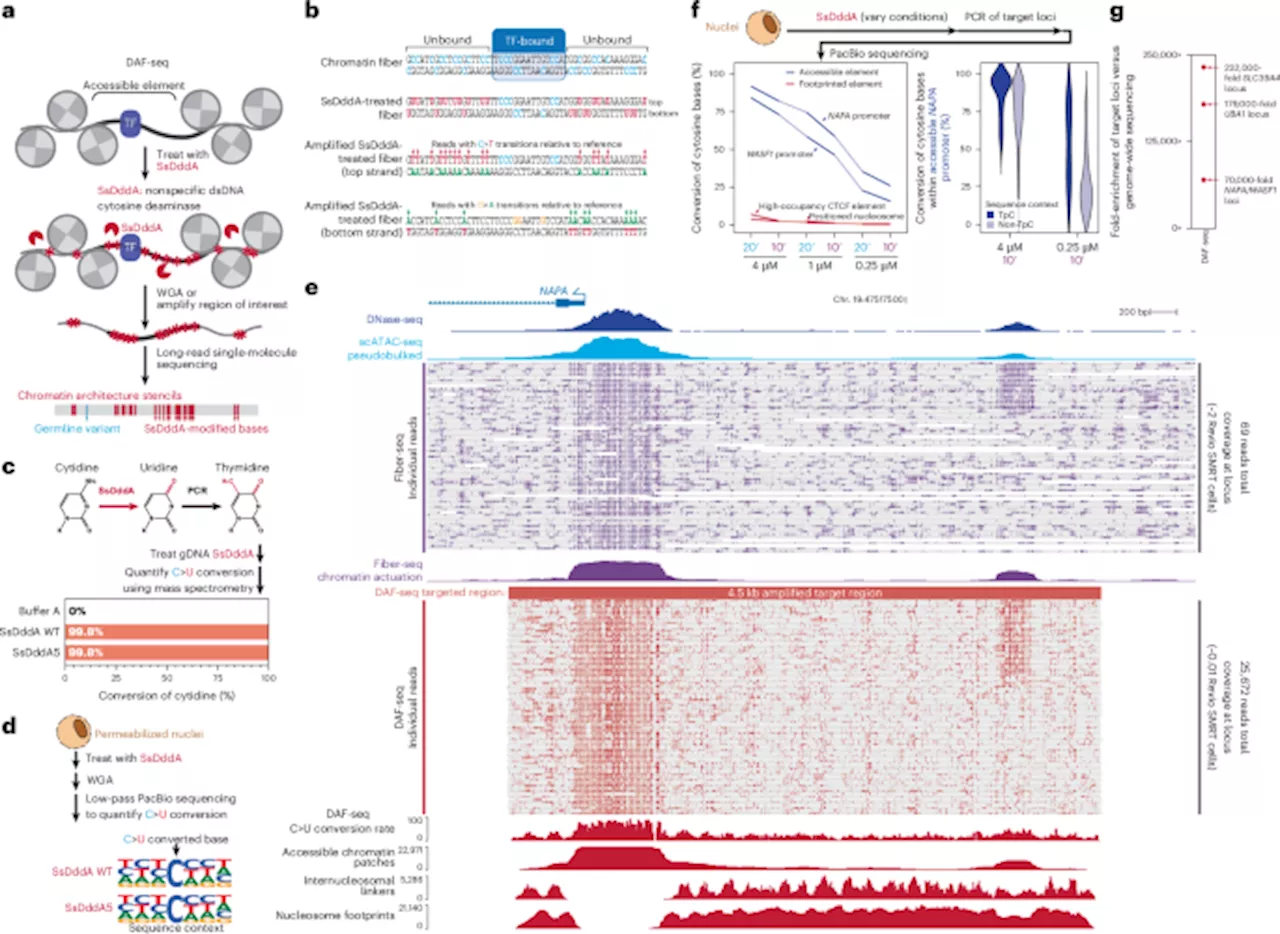
Researchers have introduced a novel method for mapping chromatin architectures at the single-cell level, enhancing our understanding of gene regulation in diploid organisms. This innovative approach, known as Deaminase-Assisted single-molecule chromatin Fiber sequencing (DAF-seq), allows scientists to examine protein occupancy along chromatin fibers with unprecedented precision.
In diploid organisms, gene regulation is significantly influenced by the binding of proteins to chromosome-length chromatin fibers. However, the variability in protein occupancy across different haplotypes and individual cells has not been fully understood. The DAF-seq technique addresses this gap by enabling researchers to perform single-molecule footprinting at nearly nucleotide-level resolution, which is crucial for mapping the complex interactions within chromatin.
Key Findings from DAF-seq
The DAF-seq method generates comprehensive maps of protein co-occupancy across approximately 99% of each cell’s mappable genome. This level of detail reveals extensive chromatin plasticity within and between diploid cells. Researchers discovered that chromatin actuation diverges by 61% between haplotypes within a single cell, and by 63% across different cells.
Further analysis indicates that regulatory elements are often co-actuated along the same chromatin fiber in a distance-dependent manner, reflecting the organization by cohesin-mediated loops. This insight is vital for understanding how regulatory elements interact to influence gene expression.
The research, conducted by a team from the University of Washington, highlights the functional significance of somatic variants and rare chromatin epialleles. With DAF-seq, scientists can now explore the intricate regulatory architectures that govern gene expression, enhancing our understanding of various genetic conditions.
Support and Collaboration
The development of DAF-seq was supported by several prestigious institutions, including the National Institutes of Health (NIH) and the Chan Zuckerberg Initiative. The research team acknowledges the contributions of various colleagues and facilities, including the Northwest Genome Center and the UW Mass Spectrometry Center, for their assistance in sequencing and mass spectrometry experiments.
A.B. Stergachis, a key researcher, holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is recognized as a Pew Biomedical Scholar. This study not only advances the field of genomics but also opens new avenues for exploring the complexities of gene regulation and its implications for health and disease.
As this research progresses, the implications for understanding genetic diversity and disease mechanisms could be significant, potentially guiding future therapeutic strategies. The full details of the study are available in the Journal of Genome Sciences, providing a comprehensive look at the methodologies and findings of this groundbreaking work.
-
 Science2 months ago
Science2 months agoOhio State Study Uncovers Brain Connectivity and Function Links
-

 Politics2 months ago
Politics2 months agoHamas Chief Stresses Disarmament Tied to Occupation’s End
-

 Science1 month ago
Science1 month agoUniversity of Hawaiʻi Joins $25.6M AI Project for Disaster Monitoring
-
 Science4 weeks ago
Science4 weeks agoALMA Discovers Companion Orbiting Giant Star π 1 Gruis
-

 Entertainment2 months ago
Entertainment2 months agoMegan Thee Stallion Exposes Alleged Online Attack by Bots
-

 Science2 months ago
Science2 months agoResearchers Challenge 200-Year-Old Physics Principle with Atomic Engines
-

 Entertainment2 months ago
Entertainment2 months agoPaloma Elsesser Shines at LA Event with Iconic Slicked-Back Bun
-

 World2 months ago
World2 months agoFDA Unveils Plan to Cut Drug Prices and Boost Biosimilars
-

 Science2 months ago
Science2 months agoInnovator Captures Light at 2 Billion Frames Per Second
-

 Business2 months ago
Business2 months agoMotley Fool Wealth Management Reduces Medtronic Holdings by 14.7%
-

 Top Stories2 months ago
Top Stories2 months agoFederal Agents Detain Driver in Addison; Protests Erupt Immediately
-

 Entertainment2 months ago
Entertainment2 months agoBeloved Artist and Community Leader Gloria Rosencrants Passes Away








