Health
Scientists Discover How OTULIN Regulates Tau Protein in Alzheimer’s

Research from the University of New Mexico Health Sciences Center and the University of Tennessee Health Science Center has revealed a critical mechanism by which the brain enzyme OTULIN regulates the expression of tau, the protein linked to the formation of toxic tangles in Alzheimer’s disease. Published in the journal Genomic Psychiatry, the study suggests that OTULIN plays a dual role, functioning not only in protein degradation pathways but also as a master regulator of gene expression and RNA metabolism.
The research team, led by Dr. Kiran Bhaskar and Dr. Francesca-Fang Liao, made this groundbreaking discovery while studying how neurons manage abnormal tau aggregates. Their unexpected findings could pave the way for new therapeutic approaches to Alzheimer’s disease and related dementias, which affect millions globally.
Dr. Bhaskar articulated the team’s initial goal, stating, “We set out to test whether stabilizing a specific type of ubiquitin chain would help clear toxic tau from neurons.” The results, however, revealed that OTULIN acts as a master switch controlling the production of tau itself.
Paradigm Shift in Understanding Tau Production
Initially, the research team hypothesized that inhibiting OTULIN’s enzymatic activity would facilitate tau clearance. Contrary to their expectations, completely knocking out the OTULIN gene in neurons resulted in the complete absence of tau—not due to enhanced degradation, but because it was not produced at all. As Dr. Liao noted, “This was a paradigm shift in our thinking.”
The study involved neurons derived from a patient with late-onset sporadic Alzheimer’s disease, which exhibited higher levels of both OTULIN protein and phosphorylated tau compared to healthy control neurons. This correlation suggested a potential link between OTULIN and disease progression.
The research yielded several significant insights:
1. In neuroblastoma cells lacking OTULIN, comprehensive RNA sequencing revealed dramatic shifts in gene expression—13,341 genes were downregulated, and 774 were upregulated, alongside alterations in RNA transcripts.
2. Pharmacological inhibition of OTULIN’s activity with a novel small molecule inhibitor, UC495, reduced levels of phosphorylated tau in Alzheimer’s neurons, indicating possible therapeutic benefits without the need for complete gene elimination.
3. The absence of OTULIN led to the upregulation of genes linked to RNA degradation and stability regulation, including components of the CCR4-NOT complex and various RNA-binding proteins associated with neurodegenerative diseases.
4. Bulk RNA sequencing of Alzheimer’s neurons revealed significant downregulation of OTULIN long noncoding RNA and decreased expression of melanoma antigen gene family members, which are involved in protein quality control.
Implications for Alzheimer’s Treatment
The implications of these findings extend beyond basic science, potentially offering new avenues for treating tauopathies, a category of neurodegenerative diseases characterized by toxic tau accumulation. Dr. Bhaskar emphasized, “OTULIN could serve as a novel drug target, but our findings suggest we need to modulate its activity carefully rather than eliminate it completely.”
The research demonstrated that partial inhibition using UC495 decreased pathological forms of tau without eliminating total tau or causing toxicity to neurons. This suggests a therapeutic window exists where OTULIN’s activity can be adjusted for beneficial effects.
Additionally, the team discovered that OTULIN deficiency prevents autoinflammation in neurons by downregulating components of inflammatory pathways, enhancing understanding of how cells maintain a balance between protein quality control and inflammatory responses.
The study also sheds light on broader RNA biology, revealing fundamental mechanisms of RNA metabolism regulation in neurons. The researchers identified upregulation of transcriptional repressors in OTULIN-deficient cells, as well as alterations in RNA-binding proteins that influence mRNA stability.
As Dr. Liao described, “We’re essentially looking at a previously unknown checkpoint in gene expression.” The study highlights connections between OTULIN and various neurodegenerative disease-associated RNA-binding proteins, suggesting implications for understanding a range of brain disorders.
The team employed advanced techniques, including CRISPR-Cas9 gene editing, induced pluripotent stem cell-derived neurons from Alzheimer’s patients, comprehensive bulk RNA sequencing, and computational drug design to identify UC495. They validated their findings across multiple cell types, bolstering the relevance to human disease.
Moving forward, the researchers aim to clarify how OTULIN influences gene expression and RNA metabolism at the molecular level. They are also investigating whether controlled OTULIN inhibition can effectively reduce tau pathology in animal models of Alzheimer’s disease.
“This discovery opens up an entirely new research direction,” stated Dr. Bhaskar. The ongoing investigations will also explore the reasons behind the reduced levels of OTULIN long noncoding RNA in Alzheimer’s neurons and whether restoring these levels could normalize OTULIN protein expression and tau pathology.
For further details, refer to the original study: “The deubiquitinase OTULIN regulates tau expression and RNA metabolism in neurons,” published in Genomic Psychiatry (2025). DOI: 10.61373/gp025a.0116.
-
 Science1 month ago
Science1 month agoOhio State Study Uncovers Brain Connectivity and Function Links
-

 Politics1 month ago
Politics1 month agoHamas Chief Stresses Disarmament Tied to Occupation’s End
-

 Entertainment1 month ago
Entertainment1 month agoMegan Thee Stallion Exposes Alleged Online Attack by Bots
-

 Science4 weeks ago
Science4 weeks agoUniversity of Hawaiʻi Joins $25.6M AI Project for Disaster Monitoring
-

 Science2 months ago
Science2 months agoResearchers Challenge 200-Year-Old Physics Principle with Atomic Engines
-

 Entertainment1 month ago
Entertainment1 month agoPaloma Elsesser Shines at LA Event with Iconic Slicked-Back Bun
-

 World1 month ago
World1 month agoFDA Unveils Plan to Cut Drug Prices and Boost Biosimilars
-

 Top Stories1 month ago
Top Stories1 month agoFederal Agents Detain Driver in Addison; Protests Erupt Immediately
-

 Business1 month ago
Business1 month agoMotley Fool Wealth Management Reduces Medtronic Holdings by 14.7%
-

 Entertainment1 month ago
Entertainment1 month agoBeloved Artist and Community Leader Gloria Rosencrants Passes Away
-

 Politics1 month ago
Politics1 month agoNHP Foundation Secures Land for 158 Affordable Apartments in Denver
-
 Science2 weeks ago
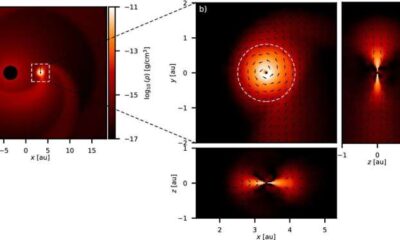
Science2 weeks agoALMA Discovers Companion Orbiting Giant Star π 1 Gruis









