Science
Tokyo University Research Breakthroughs RNA Folding with Simulations
Research from the **Tokyo University of Science** has made significant strides in understanding RNA folding through advanced molecular dynamics simulations. In a study published on **November 4, 2025**, Associate Professor **Tadashi Ando** led a comprehensive evaluation of simulation methods that could reliably model the complex folding processes of RNA molecules.
Ribonucleic acid (**RNA**) is crucial in various biological functions, extending beyond its role as a messenger of genetic information. Its ability to form intricate three-dimensional structures makes it essential in gene regulation and biotechnology, particularly as interest in RNA-based therapeutics grows. Accurately predicting these structures is vital for maximizing the potential of RNA in medical applications.
Historically, simulating RNA folding from an unfolded state has posed challenges in computational biophysics. Previous studies primarily focused on simple stem-loop structures, often yielding limited success. Dr. Ando’s research sought to address this gap by employing a combination of state-of-the-art tools in molecular dynamics simulations.
Innovative Simulation Techniques
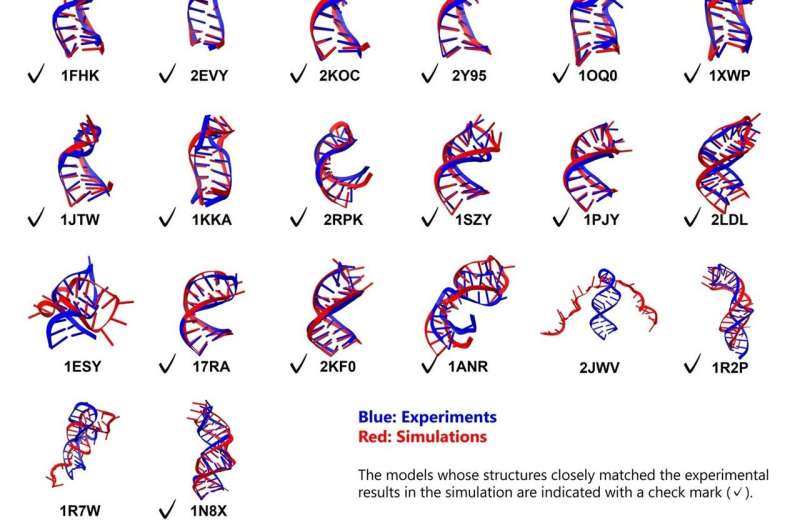
The study utilized two advanced components: the **DESRES-RNA atomistic force field** and the **GB-neck2 implicit solvent model**. The latter approximates the surrounding environment as a continuous medium, significantly accelerating the conformational sampling process compared to explicit models. This approach allowed for the modeling of a diverse range of **RNA stem-loops**, encompassing 26 different structures with lengths varying from **10 to 36 nucleotides**.
All simulations commenced from fully extended, unfolded conformations. The findings revealed a high degree of stability and accuracy in folding, validating the efficacy of the combined **DESRES-RNA** and **GB-neck2** tools. Notably, **23 out of 26 RNA molecules** successfully folded into their expected structures. For the simpler motifs, the simulations achieved exceptional accuracy, with root mean square deviation (**RMSD**) values of less than **2 Å** for stems and less than **5 Å** overall.
Even more complex structures, including five out of eight that featured bulges or internal loops, were successfully simulated. This achievement marks a notable milestone, as Dr. Ando pointed out, stating, “Previous studies have reported only two or three folding examples of simple stem-loop structures of approximately 10 residues, whereas this study deals with structures of much greater scale and complexity.”
Implications for RNA Research
The successful simulation of a broad and complex library of RNA structures represents a significant advance in computational biology. It confirms the reliability of the chosen methods for investigating large-scale conformational changes in RNA. Moreover, the study offers pathways for future refinement of simulation tools, particularly in enhancing the accuracy of loop regions, which exhibited RMSD values around **4 Å**.
As RNA becomes increasingly relevant in drug development, understanding its folding mechanisms is crucial. This knowledge could pave the way for new treatments targeting genetic disorders, viral infections such as **COVID-19**, and various cancers. As Dr. Ando concluded, “The ability to reproduce the overall folding of the basic stem-loop structure is an important milestone in understanding and predicting the structure, dynamics, and function of RNA using highly accurate and reliable computational models.”
This research not only enhances our understanding of RNA but also propels the development of more effective RNA-based medical tools. The advancements in simulation techniques could have far-reaching implications in the fields of biotechnology and pharmacology.
-

 Politics1 week ago
Politics1 week agoHamas Chief Stresses Disarmament Tied to Occupation’s End
-

 Science3 weeks ago
Science3 weeks agoResearchers Challenge 200-Year-Old Physics Principle with Atomic Engines
-
 Science1 week ago
Science1 week agoOhio State Study Uncovers Brain Connectivity and Function Links
-

 Top Stories1 week ago
Top Stories1 week agoFederal Agents Detain Driver in Addison; Protests Erupt Immediately
-

 Entertainment1 week ago
Entertainment1 week agoMegan Thee Stallion Exposes Alleged Online Attack by Bots
-

 Entertainment2 weeks ago
Entertainment2 weeks agoSyracuse Stage Delivers Lively Adaptation of ‘The 39 Steps’
-

 World3 weeks ago
World3 weeks agoGlobal Military Spending: Air Forces Ranked by Budget and Capability
-

 Top Stories1 week ago
Top Stories1 week agoWill Smith Powers Dodgers to World Series Tie with Key Homer
-

 Politics3 weeks ago
Politics3 weeks agoNHP Foundation Secures Land for 158 Affordable Apartments in Denver
-

 Top Stories1 week ago
Top Stories1 week agoOrioles Hire Craig Albernaz as New Manager Amid Rebuild
-

 Lifestyle1 week ago
Lifestyle1 week agoTrump’s Push to Censor National Parks Faces Growing Backlash
-

 Politics1 week ago
Politics1 week agoNFL Confirms Star-Studded Halftime Show for Super Bowl LVIII